Genomes of five cotton species unveiled by Texas-rich research team
Cotton sequences will provide long-needed genomics resources, insights to facilitate genetic improvements
Cotton – we touch it every day. From clothes to medical supplies to animal feed, cotton continues to increase in quality. A recent collaborative, including Texas A&M researchers, is making sure this amazing crop, and thus the products made from it, will continue to be efficiently bred, grown and produced.

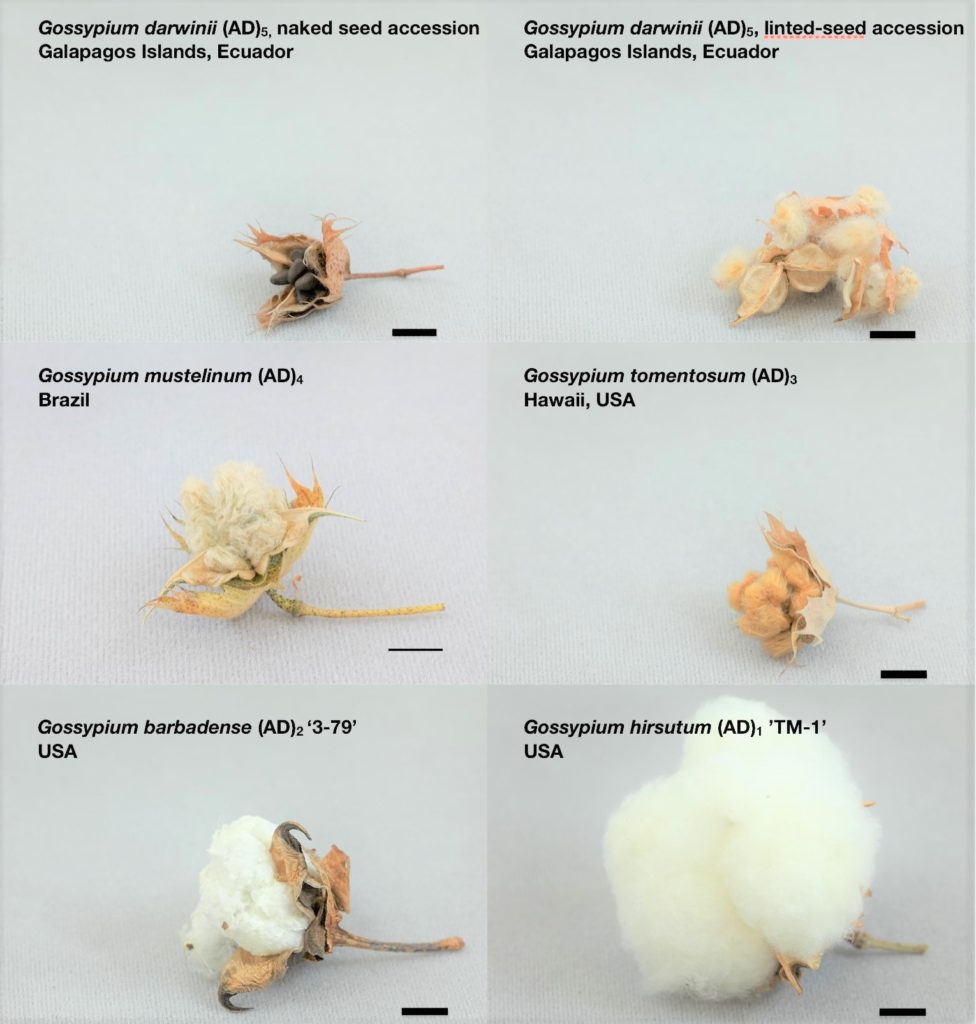
The multi-institutional research team sequenced five cotton species, including Upland and Pima cotton grown here in Texas, as well as globally. Contributions to the effort from Texas involved Texas A&M University, Texas A&M AgriLife Research and the University of Texas – Austin.
The most recent issue of Nature Genetics reports on the results of this collaboration — high-quality genome-wide sequence assemblies for each of five 52-chromosome species of the cotton genus Gossypium, a member of the Malvaceae family, which also includes okra, kenaf, hibiscus, durian and cacao.
The overall project was funded primarily by the National Science Foundation, and led by Z. Jeffrey Chen, Ph.D., a former student and former faculty member of Texas A&M who now holds the D. J. Sibley Centennial Professorship in Plant Molecular Genetics at the University of Texas at Austin.
Breeding cotton typically increases economic yield through better productivity, better quality of products and improved sustainability by providing better pest resistance and drought resilience, David Stelly, Ph.D., a co-principal investigator in the National Science Foundation project and AgriLife Research cotton breeder in the Texas A&M Department of Soil and Crop Sciences, College Station.
“Globally, cotton is the premier natural fiber crop of the world, a major oilseed crop and an important feed crop,” Stelly said. “This report establishes new opportunities in multiple basic and applied scientific disciplines that relate directly and indirectly to genetic diversity, evolution, wild germplasm utilization and increasing the efficacy with which we use natural resources for provisioning society.”

“This kind of mega-project takes a lot of time and effort, but can yield game-changing results, and this one certainly has done that, Already, we are seeing paradigm shifts in what we and others are doing and thinking about doing. These kinds of data are vital to our research and breeding efforts and open many doors for exploration.”
David Stelly, Ph.D.
Texas A&M AgriLife Research cotton breeder
The cotton genome research project
While fiber removed from the cotton seed is of greatest value, ginned seed also provides significant additional value as a source of vegetable oil and/or dairy cattle feed. The recent data and findings provide immediately accessible resources for basic and applied research, including breeding and gene editing.
The other three species sequenced originate from Hawaii, the Galapagos Islands or Ecuador and Brazil. They remain undomesticated but are sources of prospectively useful genetic differences. The Nature Genetics report should facilitate use of all five species in genomics-aided cotton breeding programs.
Stelly said the importance of the assemblies may be accentuated by the extreme complexity of cotton’s genome. It contains a relatively large number of genes, about twice as many as occur in most flowering plants with simple genomes.
The researchers report that sequences of these five species’ genomes will provide long-needed genomics resources and insights that will facilitate genetic improvements needed to maintain economic yield from production, enhance quality and value of the fiber and seed products, and further improve sustainability-enhancing features, such as resistance to pests, pathogens, drought and heat-resilience.
Contributions from Stelly’s laboratory
Contributions from Texas A&M came through Stelly’s laboratory. A key finding by graduate student Luis De Santiago was the detection and mapping of numerous “haplotypic blocks” throughout the genome of Upland cottons.
Stelly explained these present a major challenge for breeding, because they are both non-recombinant and virtually uniform among cultivars. Evidence corroborating the haplotypic blocks was obtained from analyses of genetic recombination, also involving Yu-Ming Li and former student Amanda Hulse-Kemp, Ph.D.
Also, from Stelly’s laboratory, researchers Robert Vaughn, Ph.D., provided plant, seed and nuclei acid samples to the team, and Bo Liu, Ph.D., provided integrative molecular cytogenetic mapping data.
“This kind of mega-project takes a lot of time and effort, but can yield game-changing results, and this one certainly has done that,” Stelly said. “Already, we are seeing paradigm shifts in what we and others are doing and thinking about doing. These kinds of data are vital to our research and breeding efforts and open many doors for exploration.”
He also emphasized collaborations and individual contributions are instrumental to success.
“Research projects like this unlock agriculture’s potential,” said Patrick J. Stover, vice chancellor of Texas A&M AgriLife, dean of the College of Agriculture and Life Sciences and director of Texas A&M AgriLife Research. “By developing crops that enhance health and increase profitability, we not only improve cotton immediately, but the way we approach this data and findings provide direction for basic and applied research far into the future.”
“The Soil and Crop Sciences Department appreciates the leadership of Dr. Stelly in guiding this project to completion and providing the vision for implementing the results to benefit our cotton producers,” said David Baltensperger, Ph.D., department head, College Station.
Other members of the team
Other project members include:
- Chen’s functional genomics / epigenetics team at UT-Austin.
- Jane Grimwood, Ph.D., and Jeremy Schmutz, along with their HudsonAlpha/JGI structural genomics and bioinformatics teams, including Jerry Jenkins, Ph.D., and key bioinformatics contributor Avinash Sreedasyam, Ph.D.
- The U.S. Department of Agriculture genomics and bioinformatics teams of Brian Scheffler, Ph.D., Mississippi, and Hulse-Kemp, North Carolina.
- The Clemson genomics team of Chris Saski, Ph.D.
- Keith McGee, Ph.D., and his educational team at Alcorn State.
- Mississippi State genomics group of Dan Peterson, Ph.D.
- The Iowa State taxonomic genomics group involving Jonathan Wendel and Corrinne Grover, both Ph.Ds.
- Industry involvement through Don Jones, Ph.D., with Cotton Incorporated, a not-for-profit company that works with cotton scientists, the textile industry and consumers.
Other institutions involved in the research were Nanjing Agricultural University in China, and the U.S. Department of Energy Joint Genome Institute. The work was supported by grants from the U.S. National Science Foundation, U.S. Department of Agriculture and Cotton Incorporated. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy. The work was also supported by grants from National Natural Science Foundation of China, Jiangsu Collaborative Innovation Center for Modern Crop Production, and Natural Science Foundation of Zhejiang Province, China.