Discovery in fight against drug-resistant fungal infections
Researchers discover a promising drug target to counter the rise of drug-resistant fungal ‘superbugs’
Developing new antifungal treatments is a rising health priority due to an alarming rise in multidrug-resistant fungal “superbugs” that evade medications clinicians have relied on for decades.
Now, a Texas A&M University System team’s work is launching the next steps toward the discovery of new antifungal medications.
Drug-resistant fungal species affect at least 3.6 million people annually in the U.S. alone, with a direct medical cost of $3 billion. Fungal infections can progress to severe illness or death, and the window for effective treatment is short. Hospital patients with suppressed immune systems are particularly vulnerable to these pathogens.
However, fungal cells share many similarities with human cells, so treatments effective against fungi can have undesired toxic effects on human cells. Newly developed drugs must efficiently limit fungal survivability without causing additional complications in patients.
Therefore, identifying both the right targets in fungi and the small molecule inhibitors that restrain those target activities with exquisite sensitivity are necessary first steps in antifungal drug discovery.
Both these first steps were achieved in a study recently published in the Journal of Biological Chemistry. The study’s senior authors are Tatyana Igumenova, Ph.D., associate professor in the Department of Biochemistry and Biophysics at the Texas A&M College of Agriculture and Life Sciences, and Vytas Bankaitis, Ph.D., the E.L. Wehner-Welch Chair in the Department of Cell Biology and Genetics at the Texas A&M University School of Medicine. Xiao-Ru Chen, the lead author of the work, is a graduate student in the Department of Biochemistry and Biophysics.
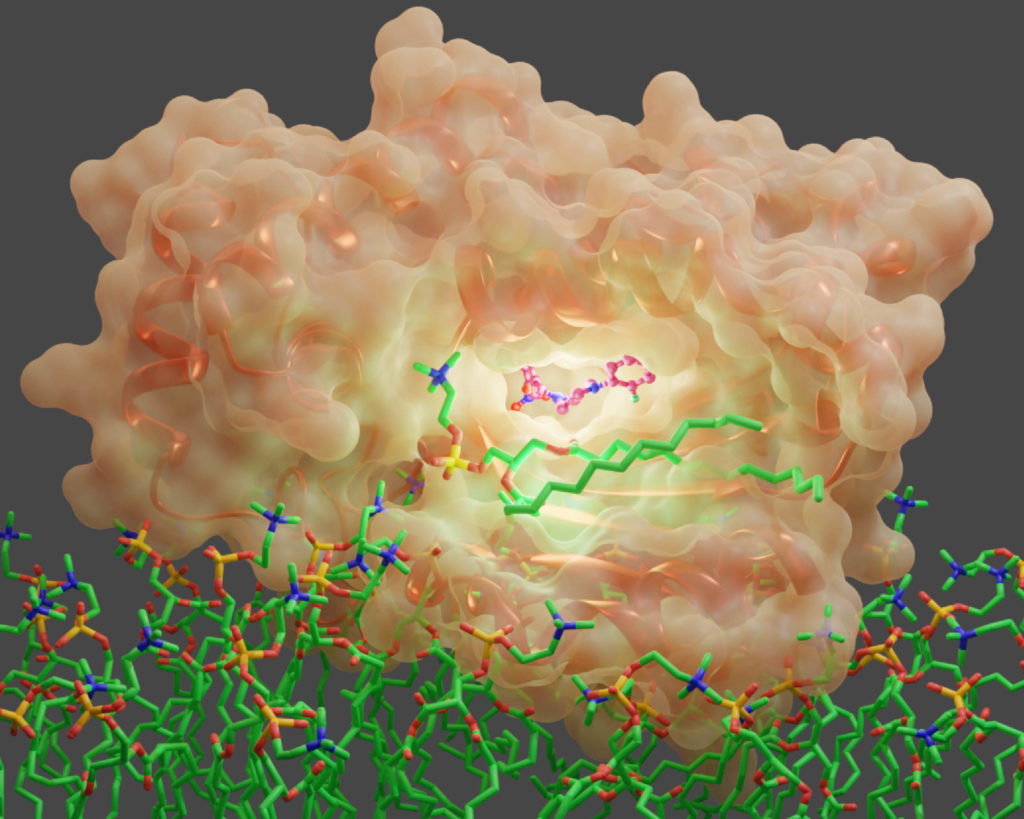
Specifically, the research team uncovered how diverse small molecules inhibit an essential regulator of a fungal signaling molecule called Sec14. Sec14 is a protein that executes membrane trafficking functions that fungal cells need to grow and form biofilms. It is also essential for fungal survival and disease development in pathogenic fungi.
Grants from the National Institutes of Health and the Robert A. Welch Foundation helped fund the research.
An example of basic and translational research
The Journal of Biological Chemistry highlighted this study as a “Recommended Read” — a designation it reserves for papers of especially high quality.
“This study is an excellent example of the tight linkage between basic science and the development of new directions of translational science,” Bankaitis said. “From the basic science perspective, the small molecule inhibitors are proving to be enormously useful as tool compounds by which we can dissect how Sec14 functions as a signaling regulator. This problem is of intense interest as Sec14 and other members of the Sec14 protein family are functionally mysterious from a biochemical and biophysical point of view. Yet that very same body of information translates directly to developing rational strategies for producing next-generation antifungal drugs.”
Toward the next steps in antifungal drug discovery
Using baker’s yeast as a model fungus, team members identified four new classes of antifungal molecules that inhibit Sec14 proteins in yeast and in some virulent fungi. These small molecule inhibitors, or SMIs, compete with native fungal ligands that Sec14 depends on to execute signaling functions. When these new inhibitors bind to Sec14, they interfere with fungal virulence.
“The application of cutting-edge integrative structural biology approaches was essential to decipher the mechanisms by which Sec14 was inhibited by each SMI,” Igumenova said. “It required the use of X-ray crystallography, molecular dynamics simulations and high-resolution nuclear magnetic resonance spectroscopy to obtain an atomistic description of how Sec14 interacts with SMIs and the principles behind the exquisite Sec14 binding specificity. Of particular note is our innovative use of fluorine nuclear magnetic resonance spectroscopy to directly monitor the competition between native fungal ligands and SMIs.”
The team’s discovery of how SMIs bind to Sec14 now launches the next steps in drug discovery. This research validates a new target and details new strategies for rational structure-based development of next-generation antifungal drugs.
Story written by Heidi Creed, graduate assistant researcher, Department of Medical Physiology, Texas A&M University School of Medicine.


